Aiming at the abiotic stresses of dry-hot wind and high temperature during the critical stages of wheat growth and development in Huang-Huai-Hai region of China, we identified and reported the total members of the whole heat shock transcription factor(Hsf) family and special subclass C3 of wheat firstly. The results showed that gene structure, biological characteristics and functions among subclass A, B and C are diverse and complex. Some subclass A2 genes contribute to improve the thousand grain weight(TGW) and decrease stress susceptibility index(SSI) of wheat under heat stress, and group C genes perhaps mainly participate in the thermotolerance and drought regulations in later development stages of wheat. The results will provide theoretical evidence for further investigation the thermotolerance mechanism of wheat.
The representative paper “Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L.” was published publicly in BMC Genomics on April 1st of 2019(Q2, IF3.501). The first author is Duan Shuonan and the correspondences are Dr. Guo Xiulin and Dr. Li Guoliang, the corresponding affiliation is Institute of Genetics and Physiology, Hebei Academy of Agriculture and Forestry Sciences/Plant Genetic Engineering Center of Hebei Province.
Abstract:
Background: Enhancement of crop productivity under various abiotic stresses is a major objective of agronomic research. Wheat (Triticum aestivum L.) as one of the world’s staple crops is highly sensitive to heat stress, which can adversely affect both yield and quality. Plant heat shock factors (Hsfs) play a crucial role in abiotic and biotic stress response and conferring stress tolerance. Thus, multifunctional Hsfs may be potentially targets in generating novel strains that have the ability to survive environments that feature a combination of stresses.
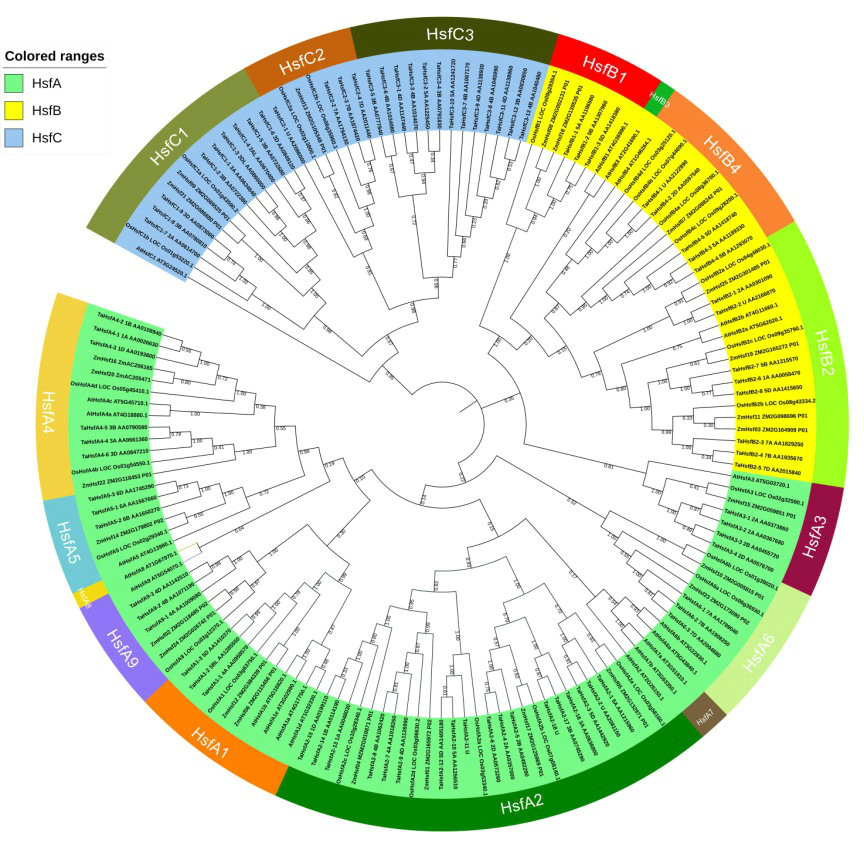
Result: In this study, using the released genome sequence of wheat and the novel Hsf protein HMM(Hidden Markov Model) model constructed with the Hsf protein sequence of model monocot(Oryza sativa) and dicot(Arabidopsis thaliana) plants, genome-wide TaHsfs identification was performed. Eighty-two non-redundant and fulllength TaHsfs were randomly located on 21 chromosomes. The structural characteristics and phylogenetic analysis with Arabidopsis thaliana, Oryza sativa and Zea mays were used to classify these genes into three major classes and further into 13 subclasses. A novel subclass, TaHsfC3 was found which had not been documented in wheat or other plants, and did not show any orthologous genes in A. thaliana, O. sativa, or Z. mays Hsf families. The observation of a high proportion of homeologous TaHsf gene groups suggests that the allopolyploid process, which occurred after the fusion of genomes, contributed to the expansion of the TaHsf family. Furthermore, TaHsfs expression profiling by RNA-seq revealed that the TaHsfs could be responsive not only to abiotic stresses but also to phytohormones. Additionally, the TaHsf family genes exhibited class-, subclass- and organ-specific expression patterns in response to various treatments.
Conclusions: A comprehensive analysis of Hsf genes was performed in wheat, which is useful for better understanding one of the most complex Hsf gene families.
Keywords: Genome-wide, Heat shock factor, Triticum aestivum, Expression profile, Abiotic stress
Corresponding author:
Guo Xiulin
Mobile Tel: +8613032602938
E-mail: myhf2002@163.com
Fig.1 Neighbor-joining phylogenetic tree of wheat, rice, Arabidopsis and maize Hsf families