On November 5, 2020, the team of Biocontrol for Plant Disease, in Plant Protection Institute, Hebei Academy of Agriculture and Forestry Sciences, published an online research article entitled "Genome mining and UHPLC – QTOF – MS / MS to identify the potential antimicrobial compounds and determine the specificity of biological gene clusters in Bacillus subtilis NCD-2" in BMC genomics. This study reported the antifungal active compounds produced by B. subtilis NCD-2, found an unique fengycin synthetic gene cluster, and revealed a potential new mechanism of fengycin biosynthesis in strain NCD-2.
Background
Bacillus subtilis NCD-2 was isolated from the cotton rhizosphere and showed strong inhibitory ability against phytopathogens. The microbial-fungicide using strain NCD-2 as the main active component had been formally registered in 2010 in China, and could successfully suppress plant soil-borne diseases. To better understand the mechanism of strain NCD-2 suppressing plant soil-borne diseases and improve its biocontrol efficiency, the main antimicrobial active compounds should be clarified. In this study, the potential antimicrobial compounds were identified by using Genome mining and Ultra-high-performance liquid chromatography coupled to quadrupole-time-of-flight tandem mass spectrometry (UHPLC-QTOF-MS/MS) technologies.
Main results
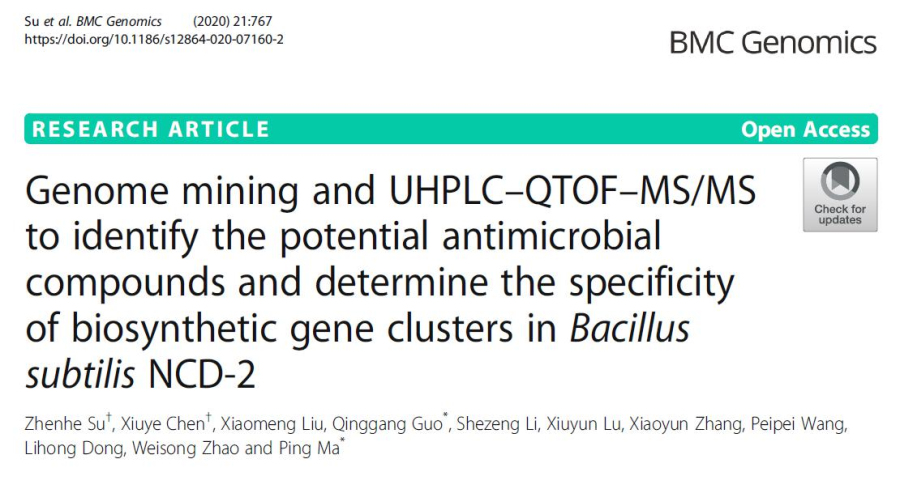
1、Nine secondary metabolite synthetic gene clusters exist in NCD-2 genome.
The antiSMASH was used to predict the secondary metabolite biosynthesis gene clusters of NCD-2 genome, and a total of nine gene clusters were predicted, involved in three NRPSs, two terpenoids, one heterozygous NRPS-PKS-other KS, one type III PKS, one sactipeptide-head to tail and one unknown product (Fig. 1). Among them, gene clusters 3, 7, 8 and 9 have 100% amino acid sequence homology with known bacillaene, bacillibactin, subtilosin and bacilysin synthetic gene clusters, respectively. The amino acid similarity between gene cluster 4 and known fengycin biosynthesis gene cluster is 93%. Compared with other fengycin producing Bacillus strains, the fengycin biosynthetic gene cluster of strain NCD-2 contains three genes fenEAB, while other strains contain five genes fenCDEAB.
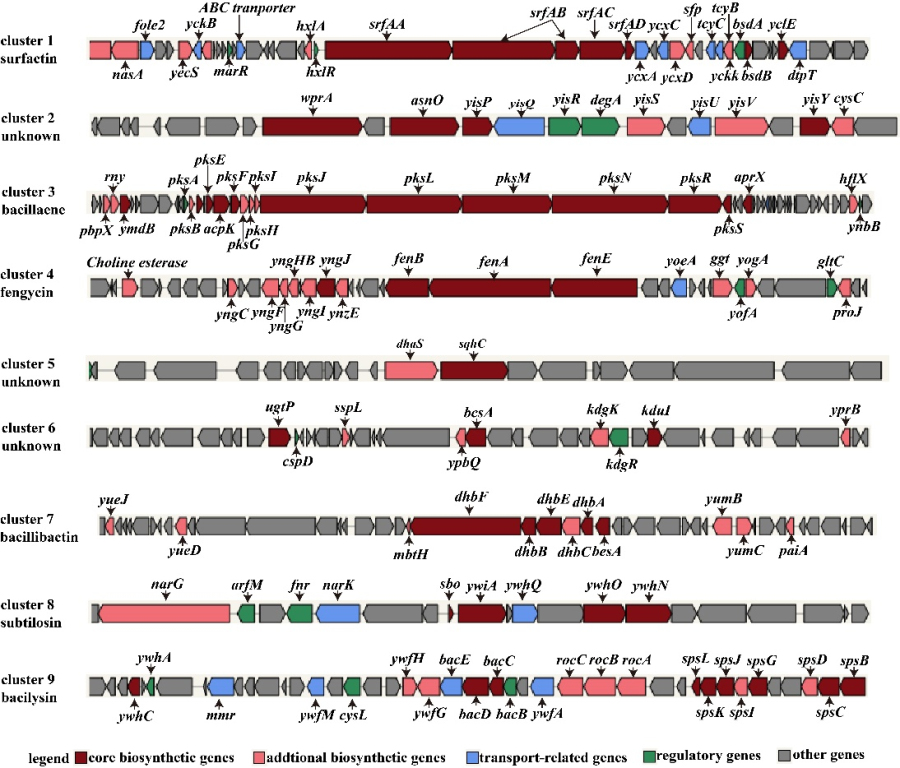
Fig. 1 Schematic diagram of nine secondary metabolite biosynthetic gene clusters of Bacillus subtilis strain NCD-2.
2、Specificity of fengycin synthetic gene cluster in strain NCD-2.
There are only three genes in the cluster of fengycin in strain NCD-2: gms1961, gms1960 and gms1959 (Fig. 2). Gms1961 corresponds to FenE in FZB42, containing conserved residues in A8 and A9 domains, binding glutamate and valine respectively (Fig. 2). Gms1960 and gms1959 correspond to FenA and FenB in FZB42, respectively. Interestingly, FenC and FenD and their homologues were not found in the genome of NCD-2. Therefore, the amino acid sequences of FenC and FenD from strain FZB42 were compared with those of strain NCD-2 by software Bioedit, and the most similar proteins were Gms1961 and Gms1960, respectively. Based on this finding, we propose the hypothesis that Gms1961 and Gms1960 play the function of FenC and FenD respectively in strain NCD-2, that is, Gms1961 of NCD-2 has FenE and FenC function, and Gms1960 has the function of FenA and FenD.
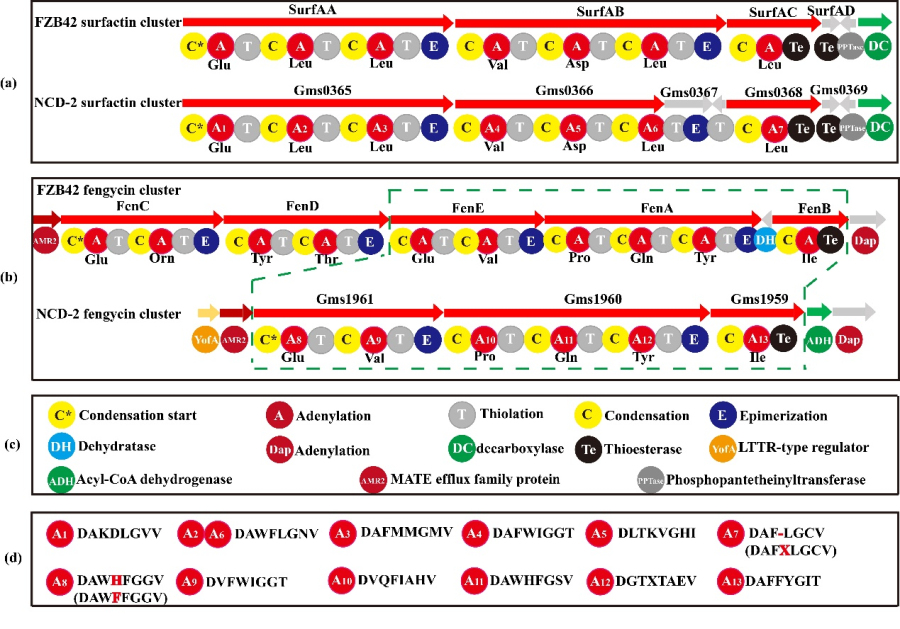
Fig. 2 Comparison of domains of core genes involved in fengycin synthesis in strain NCD-2.
3. Mass spectrometry of fengycin produced by strain NCD-2.
Fengycin was isolated from the lipopeptide extract of strain NCD-2 by FPLC. The mass spectrometry results showed that there were five components of fengycin of NCD-2 (Fig. 3), and the m/z of NCD-2 was 732.4, 746.4, 725.4, 739.4 and 767.4 (secondary mass spectrometry), corresponding to fengycin A, fengycin B, fengycin A2, fengycin B2 and fengycin C respectively. According to the length of β – hydroxyl fatty acids, five kinds of fengycin are divided into different homologues, and their chain length varies from C12 to C20.
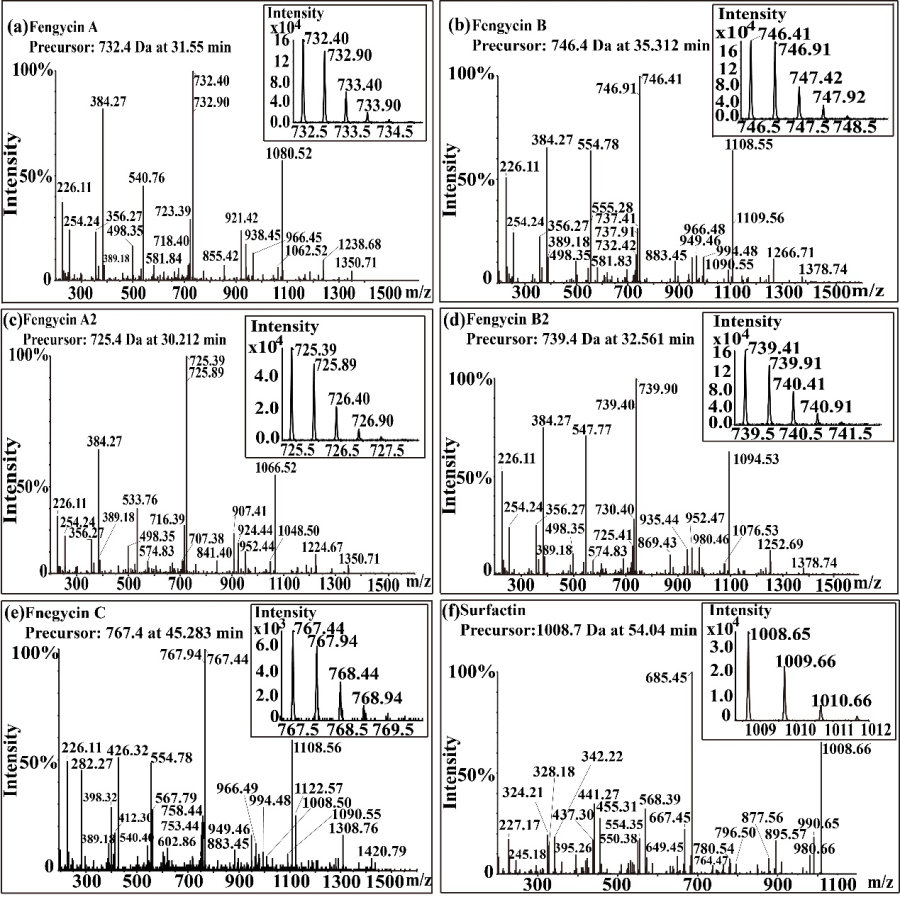
Fig. 3 Mass spectrum of fengycin.
This study not only identified bioactive compounds produced by strain NCD-2, explained the biocontrol mechanism of NCD-2 with excellent field control effect, but also for the first time found a unique fengycin synthase gene cluster, which revealed a new mechanism of fengycin synthesis by strain NCD-2, and provided a new strategy for improving the production of fengycin synthesis and the construction of engineering strain.
Article links: https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-020-07160-2